Odisha State Board CHSE Odisha Class 12 Biology Important Questions Chapter 12 Principles and Processes of Biotechnology Important Questions and Answers.
CHSE Odisha 12th Class Biology Important Questions Chapter 12 Principles and Processes of Biotechnology
Principles and Processes of Biotechnology Class 12 Important Questions CHSE Odisha
Very Short Answer Type Questions
Choose the correct option
Question 1.
GAATTC is the recognition site for which of the following restriction endonucleases?
(a) Hind III
(b) Eco RI
(c) Bam I
(d) Hae III
Answer:
(b) Eco RI
Question 2.
Given below is a sample of a portion of DNA strand giving the base sequence on the opposite strands. What is so special shown in it?
(a) Replication completed
(b) Deletion mutation
(c) Start codon at the 5′ end
(d) Palindromic sequence
Answer:
(d) Palindromic sequence
Question 3.
Agarose extracted from sea weeds is used in
(a) spectrophotometer
(b) tissue culture
(c) PCR
(d) gel electrophoresis
Answer:
(d) gel electrophoresis
Question 4.
The blotting of RNA is called
(a) Northern blot
(b) Southern blot
(c) Western blot
(d) Eastern blot
Answer:
(a) Northern blot
Question 5.
Eco RI cleaves the DNA strands to produce
(a) blunt ends
(b) sticky ends
(c) satellite ends
(d) ori replication end
Answer:
(b) sticky ends
Question 6.
DNA fragments generated by the restriction endonucleases in a chemical reaction can be separated by
(a) electrophoresis
(b) restriction mapping
(c) centrifugation
(d) polymerase chain reaction
Answer:
(a) electrophoresis
Question 7.
Commonly used vectors for human genome sequencing are
(a) T-DNA
(b) BAC and YAC
(c) expression vector
(d) T/A cloning vectors
Answer:
(b) BAC and YAC
Question 8.
Which procedure is followed for amplification of DNA?
(a) Electrophoresis
(b) Autoradiography
(c) Polymerase chain reaction
(d) Southern blotting
Answer:
(c) Polymerase chain reaction
Question 9.
The Polymerase Chain Reaction (PCR) is a technique that is used for
(a) in vivo replication of specific DNA sequence using thermostable DNA polymerase
(b) in vitro synthesis of mRNA
(c) in vitro replication of specific DNA sequence using thermostable DNA polymerase
(d) in-vivo synthesis of mRNA
Answer:
(c) In-vitro replication of specific DNA sequence using thermostable DNA polymerase.
Question 10.
The figure below shows three steps (A, B, C) of Polymerase Chain Reaction (PCR). Select the option giving correct identification together with what it represents?
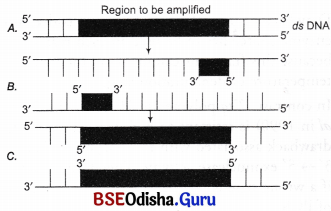
(a) B – denaturation at a temperature of about 98°C separating the two DNA strands
(b) A – denaturation at a temperature of about 50°C
(c) C – extension in the presence of heat stable DNA polymerase
(d) A – annealing with two sets of primers
Answer:
(a) B – denaturation at a temperature of about 98°C separating the two DNA strands.
Question 11.
In recombinant DNA technique, the term vector refers to
(a) plasmids that can transfer foreign DNA into a living cell
(b) cosmids that can cut DNA at specific base sequence
(c) plasmids that can join different DNA fragments
(d) cosmids that can degrade harmful proteins
Answer:
(a) plasmids that can transfer foreign DNA into a living cell
Question 12.
The rDNA molecule is introduced into the cell of bacterium with the help of.
(a) restriction endonuclease
(b) DNA ligase
(c) electroporation
(d) None of the above
Answer:
(a) restriction endonuclease
Question 13.
The bacterial source of Hpa I is
(a) Haemophilus influenzae
(b) Bacillus amyloliquefaciens
(c) Providentcia stuarth
(d) Haemophilus parainfluenzae
Answer:
(d) Haemophilus parainfluenzae
Question 14.
Klenow fragment does not possess
(a) 5′ → 3′ exonuclease
(b) 3′ → 5′ exonuclease
(c) polymerase
(d) All of the above
Answer:
(a) 5′ → 3′ exonuclease
Correct the statements, if required, by changing the underlined word(s)
Question 1.
Restriction enzymes are used to cut single-stranded DNA.
Answer:
double-stranded DNA.
Question 2.
‘CO’ part in Eco RI stands for coenzyme.
Answer:
coli.
Question 4.
The first isolated restriction endonuclease was Hind III.
Answer:
Eco RI
Question 5.
DNA ligase forms phosphodiester bonds to ligate the DNA fragments.
Answer:
It is correct
Question 6.
Plasmids present in bacterial cells are linear double helical DNA molecules.
Answer:
circular
Question 7.
Taq polymerase is used between annealing and denaturation during PCR.
Answer:
extension.
Question 8.
A hybrid of plasmid and phage is YAC.
Answer:
BAC
Question 9.
Bacteria phage are autonomously replicating circular DNA.
Answer:
Bacteriophage
Fill in the blanks
Question 1.
DNA polymerase can be obtained from ……….. .
Answer:
Thermus aquaticus
Question 2.
The usual source of restriction endonuclease used in gene cloning is ………….. .
Answer:
bacteria .
Question 3.
Other than E. coli ………….. bacteria is used in recombinant DNA technology.
Answer:
Salmonella typhimurium.
Question 4.
First letter of restriction enzymes represents …………. .
Answer:
genus
Question 5.
Alkaline phosphatase is ………. an enzyme.
Answer:
DNA ligase
Question 6.
The vector for T-DNA is ………….. .
Answer:
Agrobacterium tumefaciens.
Question 7.
In biolistic method ………….. particles coated with foreign DNA are bombarded into target cells.
Answer:
gold or tungesten.
Question 8.
……………. is most commonly used process of foreign DNA injection in animal cells.
Answer:
Microinjection
Question 9.
…………. helps in selecting transformants and eliminating non-transformants.
Answer:
Conventional method
Express in one or two word(s)
Question 1.
Molecular scissors used in recombinant technology are known as
Answer:
restriction enzymes.
Question 2.
Which enzyme helps in joining DNA fragments?
Answer:
DNA ligase.
Question 3.
Name the process of transfer of protein molecules onto a membrane.
Answer:
Western blotting
Question 4.
Single-stranded fragments are transferred onto nitrocellulose filter paper by which process?
Answer:
Northern blotting
Question 5.
Name the bacterium that yields thermostable DNA polymerases.
Answer:
Thermus aquaticus
Question 6.
What is particle gun?
Answer:
It is a technique of bombarding microparticles of gold or tungsten with foreign DNA into target cell with high velocity.
Question 7.
Name one chemical that helps foreign DNA to enter host cell.
Answer:
Transformation
Short Answer Type Questions
Question 1.
Write a short note on genetic engineering.
Answer:
Genetic engineering is a modification, of chemical nature of genetic material (DNA/RNA) and their introduction into another organism to change the phenotypic characters of that organism.
This involves recombinant nucleic acid (DNA or RNA) techniques to form new combinations of heritable genetic material followed by the incorporation of material indirecdy through vectors or directly through microinjection and other techniques.
Question 2.
What are restriction enzymes? Mention their functions in recombinant DNA technology.
Answer:
The enzymes used for cutting the DNA during recombinant DNA technology are called restriction enzymes.
Restriction enzymes function as chemical knives or molecular scissors in genetic engineering. It recognises specific nucleotide sequence and makes cuts.
Question 3.
(i) Mention the difference in the mode of action of exonuclease and endonuclease.
(ii) How does restriction endonuclease function?
Answer:
(i) Exonucleases cleave base pairs of DNA at their terminal ends (either 5′ or 3 0 while, the endonucleases cleave DNA at any point within DNA segment at specific position except terminal ends.
(ii) Restriction endonuclease Eco RI cuts the DNA strands a little away from the palindromic sequences, but between the same two bases on the two strands
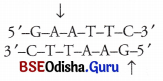
Question 4.
How are ‘sticky ends’ formed on a DNA strand? Why are they so called?
Or
A recombinant DNA is formed when sticky ends of the vector DNA and the foreign DNA join. Explain how sticky ends are formed and get joined?
Answer:
Restriction enzymes cut the strands of the DNA, a little away from the centre of the palindromic sites, but between the same two bases on opposite strands. This leaves sticky single-stranded position at the ends. These overhanging stretches are called ‘sticky ends’. These are named so, because they form hydrogen bonds with their complementary cut counterparts.
Question 5.
What does Hind and ‘III’ refer to in the enzyme Hind III?
Answer:
- The first letter ‘H indicates the genus of the organism, from which the enzyme was isolated, i.e. H-genus, Haemophilus.
- The roman number (III) denotes the sequence in which the restriction endonuclease from that particular genus, species and strain of bacteria have been isolated, i. e., third restriction endonuclease to be isolated from this species.
Question 6.
Collect five examples of palindromic DNA sequences by consulting your teacher. Better try to create a palindromic sequence by following base pair rules.
Answer:

Question 7.
Restriction enzymes should not have more than one site of action in the cloning site of a vector. Comment.
Answer:
Enzymes should not have more than one site of action.
This is because vectors have very few recognition sites for commonly used restriction enzymes.
If it will have more than one restriction sites it will generate various fragments. This will complicate the process of genetic engineering.
Question 8.
Draw agarose gel electrophoresis apparatus. Description is not required.
Answer:
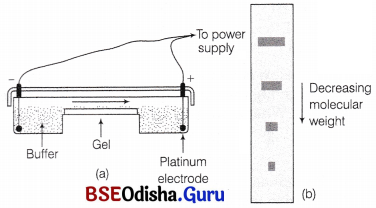
(a) Agarose gel apparatus, (b) Fluorescent bands on the agarose gel slab containing resolved DNA fragments with differing molecular weights.
Question 9.
A mixture of fragmented DNA was electrophoresed in an agarose gel. After staining the gel with ethidium bromide, no DNA bands were observed. What could be the reason?
Answer:
DNA bands may not be observed when a mixture of fragmented DNA was electrophoresed, due to following reasons
- Concentration of agarose in the gel was not proper, as greater the concentration of agarose gel used, the greater will be separation of small DNA fragments, whereas smaller the concentration of agarose, higher will be the resolution of bands.
- If the concentration of salt in the buffer was not proper.
- If DNA sample is contaminated with RNA or any other impurity or if the concentration of DNA is too low.
Question 10.
Name and describe the technique that helps in separating the DNA fragments by the use of restriction endonuclease.
Or
(i) Name the technique used for the separation of DNA fragments.
(ii) Write the type of matrix used in this technique.
(iii) How is the prepared DNA visualised and extracted for use in recombinant technology?
Answer:
(i) DNA fragments can be separated by the technique called gel electrophoresis.
(ii) The most commonly used matrix is agarose gel, which is a natural polymer extracted from sea weeds.
(iii) A compound called Ethidium Bromide (EtBr) stains DNA, which on exposure with ultraviolet radiationals gives orange light. Hence, DNA fragments appear as orange bands.
Question 11.
A plasmid DNA and a linear DNA (both of the same size) have one site for a restriction endonuclease. When cut and separated on agarose gel electrophoresis, plasmid shows one DNA band while, linear DNA shows two fragments. Explain.
Answer:
This is because plasmid is a circular DNA molecule.
When cut with enzyme, it becomes linear, but does not get fragmented. Whereas, a linear DNA molecule gets cut into two fragments. Hence, a single DNA band is observed for plasmid while, two DNA bands are observed for linear DNA in agarose gel.
Question 12.
Write the role of ‘ori’ and ‘restriction site’ in a cloning vector pBR322.
Answer:
Ori is the site where replication starts. This site is responsible for controlling the copy number of a vector. Restriction site is the site of ligation of alien/foreign DNA in the vector, in one of the two antibiotic resistance sites or coding sequence of β- galactosidase.
Question 13.
Write a short note on plasmids.
Answer:
These are small and double-stranded extrachromosomal DNA molecules having an origin of replication. They occur naturally in bacteria. Naturally occurring plasmids are not suitable for the cloning of genes. They are genetically engineered and made suitable for cloning.
Such plasmids are called cloning plasmids. A suitable cloning plasmid should preferably have the following properties
- It should be non-conjugative.
- It should have a relaxed replication and a high copy number.
- It should have an origin of replication.
- It should have unique restriction sites.
- It should have antibiotic marker gene for selection.
The most common cloning plasmid is designated as pBR322, where p stands for the word plasmid and B and R signify the names of its engineers (B. Boliver and R. Rodriguez) 322 is a numerical designation that has a relevance to these workers, who worked out the plasmid.
Question 14.
You have chosen a plasmid as vector for cloning your gene. However, this vector plasmid lacks a selectable marker. How would it affect your experiment?
Answer:
In a gene cloning experiment, first a recombinant DNA molecule is constructed, where the gene of interest is ligated to the vector and introduced inside the host cell (transformation). Since, not all the cells gets transformed with the recombinant/plasmid DNA, in the absence of selectable marker, it will be difficult to distinguish between transformants and non-transformants. Because the role of selectable marker is in the selection of transformants.
Question 15.
Draw a schematic sketch of pBR322 plasmid and label the following in it.
(i) Any two restriction sites
(ii) Ori and rop genes
(iii) An antibiotic resistant gene
Answer:
The labelled diagram of pBR322 plasmid is shown in the figure with
(i) Eco RI and Bam HI as restriction enzymes
(ii) Ori and rop genes
(iii) ampR (an antibiotic resistant gene)
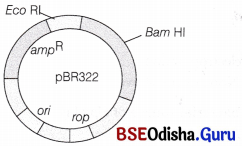
E. coli cloning vector pBR322
Question 16.
(i) How are recombinant vectors created?
(ii) For creating one recombinant vector only one type of restriction endonuclease is required. Give reason.
Answer:
(i) The vector DNA is cut at a particular restriction site using a restriction enzyme (to cut the desired DNA segment). The alien DNA is then linked with the plasmid DNA using an enzyme called ligase to form the recombinant vector.
(ii) Since, a restriction enzyme recognises and cuts the DNA at a particular sequence is called recognition site, the same restriction enzyme is used for cutting the DNA segment from both the vector and the other source to achieve as ends.
Question 17.
How bacterial cells are made competent to take up DNA?
Or
Describe the role of CaCl2 in the prepration of competent cell.
Answer:
Since, DNA is a hydrophilic molecule, it cannot pass through the cell membranes. In order to force bacteria to take up the plasmid, the bacterial cells must first be made competent to take up foreign DNA. This is done by treating them with a specific concentration of a divalent cation such as calcium (Ca<sup>2+</sup>), which increases the efficiency with which DNA enters the bacterium through the pores in its cell wall.
Recombinant DNA can then be forced into such cells by incubating the cells with recombinant DNA on ice, followed by placing them briefly at 42°C (heat shock), and then putting them back on ice. This enables the bacteria to take up the recombinant DNA.
Question 18.
PCR is a useful tool for early diagnosis of an infectious disease. Comment.
Answer:
PCR is a very sensitive technique, which enables the specific amplification of desired DNA from a limited amount of DNA containing sample. Hence, it can detect the presence of an infectious organism in the patient at an early stage of infection (even before the infectious organism has multiplied to large number).
Question 19.
(i) Explain how to find whether an E coli bacterium has transformed or not, when a recombinant DNA bearing ampicillin-resistance gene is transferred into it
(ii) What does the ampicillin-resistant gene act as, in the above case?
Answer:
(i) The recombinant/transformant can be selected out from the non-recombinants/ non-transformants by plating the transformants on ampicillin-containing medium. The transformants will grow in it, while the non-transformants will not grow.
(ii) It acts as a selectable marker.
Question 20.
Name the source of the DNA polymerase used in PCR technique. Mention, why it is used ?
Or
Give the name of the organism from where the thermostable DNA polymerase is isolated. State its role in genetic engineering.
Answer:
Bacterium Thermus aquaticus is a source of enzyme Taq polymerase. As, it is a thermostable enzyme and works at high temperature, it is used to amplify DNA in vitro by PCR. The amplified fragment of desired DNA can be used to ligate with the vector for further cloning.
Question 21.
Rearrange the following in the correct sequence to accomplish an important biotechnological reaction
(a) In vitro synthesis of copies of DNA of interest
(b) Chemically synthesised oligonucleotides
(c) Enzyme DNA polymerase
(d) Complementary region of DNA
(e) Genomic DNA template
(f) Nucleotides provided
(g) Primers
(h) Thermostable DNA-polymers (From Thermus aquaticus)
(i) Denaturation of dsDNA
Answer:
(i) Denaturation of dsDNA
↓
(e) Genomic DNA template
↓
(g) Primers
↓
(b) Chemically synthesised oligonucleotides
↓
(d) Complementary region of DNA
↓
(f) Nucleotides provided
↓
(c) Enzyme DNA polymerase
↓
(h) Thermostable DNA polymerase (from Thermus aquaticus )
↓
(a) In vitro synthesis of copies of DNA of interest
Question 22.
While doing a PCR, denaturation step is missed. What will be its effect on the process?
Answer:
If denaturation of double-stranded DNA does not take place, then primers will not be able to anneal to the template. Amplification will not occur and no extension will take place.
Question 23.
A schematic representation of PCR up to the extension stage is given below. Give answers of the following questions.
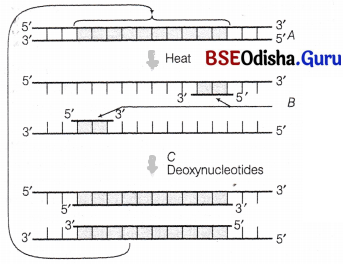
(i) Name the process A
(ii) Identify B.
(iii) Identify C and mention its importance in PCR.
Answer:
(i) A – Denaturation process
(ii) B – Primers
(iii) C – Taq DNA polymerase.
Taq polymerase is a thermostable enzyme, which remains active during the high temperature and induces denaturation of DNA.
Differentiate between the following (for complete chapter)
Question 1.
Exonucleases and Endonucleases.
Answer:
Differences between exonucleases and endonucleases are as follows
| Exonucleases | Endonucleases |
| Remove nucleotide from the outer ends of the DNA. | Make cuts at specific positions within the DNA. |
| It makes cut at only one of the two strands of DNA. | It makes cuts at both strands of DNA simultaneously. |
| e.g., snake venom and spleen phosphodiesterase. | e.g., deoxyribonuclease 1 and II. |
Question 2.
Plasmid and Chromosomal DNA.
Answer:
Differences between plasmid and chromosomal DNA are as follows
| Plasmid DNA | Chromosomal DNA |
| Extrachromosomal circular DNA. | Generally linear. |
| Not associated with histone proteins. | Associated with histone proteins. |
| Contains very few genes, but may not be necessary for the cell. | It Consists of complete genome vital for the cellular functions. |
| Replicates independently. | Replicates with genome. |
Question 3.
Electroporation and Microinjection.
Answer:
Differences between electroporation and microinjection are as follows
| Electroporation | Microinjection |
| In this method, electric field is applied to cells in order to increase the permeability of the cell membrane, allowing DNA to be introduced into the cell. | This is a vectorless method of direct delivery of recombinant DNA into plant and animal cells. |
| In this, brief pulses of high voltage electric currents are passed through the medium. | The foreign DNA is delivered into the nucleus with the help of microinjection or micropipette. |
| This procedure is mostly used for transforming plant protoplasts. | This method is particularly used for mammalian cells. |
Question 4.
Type-I endonuclease and Type II endonuclease.
Answer:
Differences between type-I endonuclease and type-II endonuclease are as follows
| Type-I endonuclease | Type-II |
| They consist of 3 different subunits. | They are structurally simple. |
| Require ATP, Mg2+, S-adenosyl-methionine for restriction. | Required Mg2+; for restriction. |
| Recognise specific sequences, but do not cut these sites. | Recognise specific sites and cut them. |
