Odisha State Board CHSE Odisha Class 12 Biology Solutions Chapter 12 Principles and Processes of Biotechnology Textbook Questions and Answers.
CHSE Odisha 12th Class Biology Chapter 12 Question Answer Principles and Processes of Biotechnology
Principles and Processes of Biotechnology Class 12 Questions and Answers CHSE Odisha
Very Short Answer Type Questions
Multiple choice questions
Question 1.
The double helical structure of DNA was proposed by
(a) Jacob and Monod
(b) Sanger and Gilbert
(c) Watson and Crick
(d) Beadle and Tatum
Answer:
(c) Watson and Crick
Question 2.
Polymerase chain reaction was discovered by
(a) H G Khorana
(b) K Mullis
(c) R Holley
(d) M Nirenberg
Answer:
(b) K Mullis
Question 3.
Exonuclease is an enzyme that
(a) makes internal cuts in polynucleotide
(b) polymerises nucleotides
(c) joins two polynucleotide fragments
(d) removes nucleotides from the termini one after another
Answer:
(d) removes nucleotides from the termini one after another
Question 4.
DNA ligase is commonly known as
(a) molecular scissors
(b) molecular marker
(c) molecular glue
(d) molecular probe
Answer:
(c) molecular glue
Question 5.
During electrophoresis, DNA fragments move from
(a) anode to cathode
(b) remain static
(c) move randomly
(d) cathode to anode
Answer:
(d) cathode to anode
![]()
Question 6.
The blotting of protein molecules to a nylon membrane is known as
(a) Southern blotting
(b) Western blotting
(c) Northern blotting
(d) Eastern blotting
Answer:
(b) Western blotting
Question 7.
Detection of a desired DNA fragment by using radioactive emission is known as
(a) hybridisation
(b) denaturation
(c) autoradiography
(d) electrophoresis
Answer:
(c) autoradiography
Question 8.
Choose the incorrect statement.
(a) A plasmid is small, double-stranded circular DNA
(b) A plasmid contains an origin of replication
(c) A plasmid has several restriction sites
(d) A plasmid has telomeres
Answer:
(d) A plasmid has telomeres
Question 9.
A cosmid is a
(a) plasmid phage hybrid vector
(b) DNA bacteriophage vector
(c) expression vector
(d) viral vector
Answer:
(a) plasmid phage hybrid vector
Question 10.
The example of a plant cell compatible vector is
(a) fertility plasmid
(b) colicinogenic plasmid
(c) tumour inducing plasmid
(d) resistance plasmid
Answer:
(c) tumour inducing plasmid
Question 11.
Amplification of DNA by PCR uses a DNA polymerase called
(a) Taq DNA polymerase
(b) RNA polymerase
(c) DNA polymerase-III
(d) Reverse transcriptase
Answer:
(a) Taq DNA polymerase
Fill in the blanks
Question 1.
The phenomenon of fermentation was demonstrated by ……………
Answer:
Louis Pasteur
Question 2.
The word ‘biotechnology’ was coined by …………..
Answer:
Karl Ereky
Question 3.
Class II restriction endonucleases (enzymes) recognise specific nucleotide sequence in DNA called ……………
Answer:
palindromic sequence
Question 4.
Cohesive ends in the DNA fragments are generated by …………… cutting.
Answer:
staggered
Question 5.
The anionic detergent, used in polyacrylamide gel electrophoresis is known as
Answer:
Sodium Dodecyl Sulphate (SDS)
Question 6.
The transfer of separated protein molecules from the gel into a nylon membrane is known as ………….
Answer:
Western blotting
Question 7.
Detection of desired DNA fragment by the emission of ionising radiation is known as ……….
Answer:
autoradiography
![]()
Question 8.
The conjoint structure formed by the joining of the vector DNA and the target DNA fragment is known as …………..
Answer:
recombinant DNA
Question 9.
The uptake of the recombinant DNA by the bacterial host cell is known as ………….
Answer:
transformation
Question 10.
The delivery of a foreign DNA fragment into the fertilised egg with a micropipette is known as …………
Answer:
microinjection
Express in one or two word(s)
1. The technique of separation of DNA fragments based on their molecular weight and electrical charge.
Answer:
Electrophoresis
2. The restriction endonuclease isolated from Escherichia coli.
Answer:
Eco RI
3. The DNA digesting enzyme that removes nucleotides from the termini.
Answer:
Exonucleases
4. The enzyme that catalyses the synthesis of RNA on a DNA template.
Answer:
RNA polymerases
5. The enzyme that catalyses the replication of DNA.
Answer:
DNA polymerases
6. The enzyme that catalyses the synthesis of a complementary DNA strand on an RNA template.
Answer:
Reverse transcriptase
7. The fluorescent dye used in agarose gel electrophoresis.
Answer:
Ethidium bromide
8. Transfer of DNA fragments from the agarose gel to a nylon membrane.
Answer:
Southern blotting
![]()
9. Breaking of hydrogen bonds in a duplex so as to make it single-stranded.
Answer:
Denaturation
10. The DNA that helps carry the target DNA fragment to the host cell for cloning.
Answer:
Vector
11. The plasmid phage hybrid cloning vector.
Answer:
Cosmid
12. A plant cell, whose cellulose cell wall is digested.
Answer:
Protoplast
13. The plasmid present in Agrobacterium tumefaciens.
Answer:
Tumour inducing (Ti) plasmid
14. Transfer of a DNA fragment into a host cell in a medium by passing brief pulses of electric current through the medium.
Answer:
Electroporation
15. The instrument used in PCR amplification.
Answer:
Thermocycler
Match the words of group ‘A with those of group ‘B’ to make meaningful pairs
Question 1.
| Group-A | Group-B |
| Restriction endonuclease | End modifying enzyme |
| DNA ligase | RNA dependent DNA polymerase |
| Exonuclease | Molecular scissors |
| Reverse transcriptase | Removes nucleotides from both ends |
| RNA polymerase | DNA dependent DNA synthesis |
| DNA polymerase | Molecular glue |
| Alkaline phosphatase | DNA dependent RNA synthesis |
Answer:
| Group-A | Group-B |
| Restriction endonuclease | Molecular scissors |
| DNA ligase | Molecular glue |
| Exonuclease | Removes nucleotides from both ends |
| Reverse transcriptase | RNA dependent DNA polymerase |
| RNA polymerase | DNA dependent RNA synthesis |
| DNA polymerase | DNA dependent DNA synthesis |
| Alkaline phosphatase | End modifying enzymes |
![]()
Question 2.
| Group-A | Group-B |
| Polyacrylamide | Agrobacterium tumefaciens |
| Southern blotting | Thermocycler |
| Plasmid | Protein electrophoresis |
| Agarose gel | Denaturation |
| Cos | Taq polymerase |
| Tumour inducing plasmid | Cloning vehicle |
| Recombinant DNA | Molecular marker |
| DNA coated tungsten particles | Nucleic acid electrophoresis |
| insertional inactivation | Chimeric DNA |
| Breaking of interchain hydrogen bonds | Particle gun |
| Equipment for PCR amplification | Screening of clones |
| Ampicillin resistance gene | Blotting of DNA |
| Thermophilus aquaticus | Cohesive site |
Answer:
| Group-A | Group-B |
| Polyacrylamide | Protein electrophoresis |
| Southern blotting | Nucleic acid electrophoresis |
| Plasmid | Cloning vehicle |
| Agarose gel | Blotting of DNA |
| Cos | Cohesive site |
| Tumour inducing plasmid | Agrobacterium tumetaciens |
| Recombinant DNA | Chimeric DNA |
| DNA coated tungsten particles | Particle gun |
| insertional inactivation | Screening of clones |
| Breaking of interchain hydrogen bonds | Denaturation |
| Equipment for PCR amplification | Thermocycler |
| Ampicillin resistance gene | Molecular marker |
| Thermophilus aquaticus | Taq polymerase |
Short Answer Type Questions
Answer each of the following in 50 words
Question 1.
Define biotechnology.
Answer:
‘Biotechnology is an application of knowledge and technique of biochemistry, microbiology, genetics, immunology, tissue and cell culture, molecular biology, chemical engineering and computer science to living systems or parts therefore, for harvesting beneficial products and/or services for mankind.
Question 2.
Define gene cloning.
Answer:
The process of making a number of identical copies of a beneficial gene is known as gene cloning. Microorganisms, especially bacteria are chosen as host cells for this work, which provide a suitable environment for replication (amplification) of genes.
The gene is introduced into a bacterium by a process, known as transformation. For transformation, a gene is delivered into a bacterial host conjointly with a carrier, also DNA, known as a vector.
Question 3.
What is a restriction endonuclease (restriction enzyme)? Why is the word restriction used to designate these?
Answer:
These enzymes recognise specific recognition sequences on DNA and make the cut either within that sequence only or at a variable distance from that sequence. The site where they cleave or cut the DNA is called recognition site.
In rDNA technology, class-II REs are used.
Question 4.
Describe two types of cutting of DNA, executed by restriction endonucleases.
Answer:
Restriction Endonucleases
Each restriction endonuclease functions by ‘inspecting’ the length of a DNA sequence.
Once, it finds its specific recognition sequence, it will bind to the DNA and cut each of the two strands of the double helix at specific points in their sugar phosphate backbones.
Each restriction endonuclease recognises a specific palindromic nucleotide sequences in the DNA. Palindromes are group of letters that form the same words when read both forward and backward, e.g., ‘MALAYALAM’. Therefore, the palindrome in DNA is a base pair sequence that is the same when read forward or background, e.g., the following sequences read the same on the two strands in 5′ → 3′ direction as well as 3′ → 5′ direction.
5′- GAATTC – 3′
3′ – CTTAAG – 5′
Restriction enzymes cut the strand of DNA a little away from the centre of the palindrome sites, but between the same two bases on the opposite strands.
![]()
Question 5.
What is electrophoresis? How many types of electrophoresis you have studied?
Answer:
The cutting of DNA by restriction endonucleases results in the generation of DNA fragments which are then separated by a technique known as gel electrophoresis.
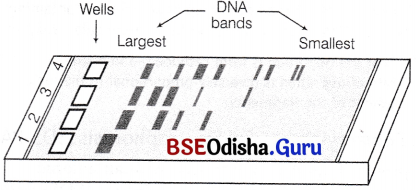
The DNA fragments are resolved (separate) according to their size through sieving effect provided by the gel when the potential difference is applied.
Two types of gels used in molecular separation are
- Agarose Used for nucleic acids (RNA and DNA).
- Polyacrylamide Used for proteins.
Question 6.
What is a palindrome? Give an example.
Answer:
Each restriction endonuclease recognises a specific palindromic nucleotide sequences in the DNA. Palindromes are group of letters that form the same words when read both forward and backward, e.gy‘MALAYALAM’. Therefore, the palindrome in DNA is a base pair sequence that is the same when read forward or background, e.g., the following sequences read the same on the two strands in 5′ → 3′ direction as well as 3′ →5′ direction.
5′- GAATTC – 3′
3′ – CTTAAG – 5′
Restriction enzymes cut the strand of DNA a little away from the centre of the palindrome sites, but between the same two bases on the opposite strands.
Question 7.
What is a polymerase? How-many types of polymerases you have studied?
Answer:
Polymerases
These enzymes catalyse the process of copying the nucleic acid molecules. These are of following types
(i) RNA polymerase These are DNA dependent RNA polymerase.
(ii) DNA polymerase They copy DNA strand into another complementary strand through replication. These are also DNA dependent enzymes.
(iii) Reverse transcriptase They catalyse the synthesis of complementary DNA strand on RNA template. These are found in retroviruses and are RNA dependent DNA polymerase.
(iv) RNA polymerase They catalyse the process of strand copying of DNA into RNA through transcription.
Question 8.
Why is DNA ligase called molecular glue?
Answer:
DNA Ligases
These enzymes help to join or seal 3′-OH end of one nucleic acid fragment with the 5′-P end of another nucleic acid fragment by forming a phosphodiester bond. As they are able to stick DNA fragments together, they are known as molecular glue. The widely used ligase enzyme is T4 DNA ligase. It is purified from E. coli cells that are infected by T4 bacteriophages.
Question 9.
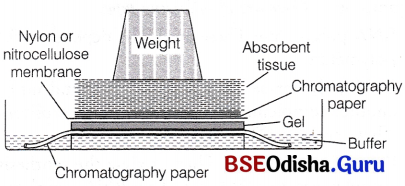
What is Southern blotting?
Answer:
Southern Blotting:
When the blotting is applied to DNA it is known as Southern blotting (named so after its discover EM Southern in 1975).
Following this, complementary DNA or RNA sequence (probe) labelled with 32 P (radioactive phosphorus) are, hybridised.
The probe binds its complementary target DNA sequence, by forming hydrogen bonds and a duplex is formed. This process is known as molecular hybridisation. The procedure that combines Southern blotting and molecular hybridisation is known as Southern blot hybridisation. After this, the target DNA sequence is identified using autoradiography.
![]()
Question 10.
Why is SDS used in polyacrylamide gel electrophoresis?
Answer:
A protein molecule contains many amino acids and all the amino acids do not have similar acid-base properties. Hence, a protein molecule cannot be conferred with uniform negative charges in a buffer solution.
It is therefore, treated with Sodium Dodecyl Sulphate (SDS), an anionic detergent that coats a protein molecule and confers negative charges uniformly.
Question 11.
What is autoradiography?
Answer:
Autoradiography:
The separated fragments in the gel are treated with an alkali to render the double-stranded fragments into single-stranded. This phenomenon is known as denaturation. These single-stranded fragments are then transferred onto a nitrocellulose filter paper or nylon membrane by a process known as blotting.
Question 12.
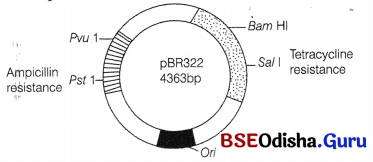
Enumerate the features of a suitable cloning plasmid.
Answer:
Plasmids:
These are small, autonomously replicating usually circular, extrachromosomal double-stranded DNA molecules that occur in many bacteria and some yeasts, but naturally occurring plasmids are not suitable for gene cloning. Therefore, they are genetically engineered so to make them compatible for cloning. These are then called as cloning vectors and, they possess the following additional properties other than these found in an ideal vector
- They are non-conjugative.
- They have relaxed replication and a high copy number.
- They should be able to clone a gene in the range of 5-10Kb
The most commonly used cloning vector is pBR322 where p = plasmid; BR = Name of its engineers
[B = Boliver and R= Rodriguez]; 322 = Number of relevant workers who worked out this plasmid. pBR322 has the following features
• It is small, ds circular DNA molecule having ori.
• It has two antibiotic marker genes namely, ampicillin resistance gene ampR and tetracyclin resistance gene (tetR). In between these two genes, Eco RI restriction site is present.
• ampR gene contains Pvu I and Pst I restriction sites.
• tetR gene contains Bam HI and Sal I restriction sites.
• The target DNA and the vector DNA fragments should be compatible for a successful RDT process.
![]()
Question 13.
What is a recombinant DNA?
Answer:
The heterogeneous combination of two different DNAs in which the gene of interest is present is known as recombinant DNA or chimeric DNA and the technology is called recombinant DNA technology.
Question 14.
What is microinjection?
Answer:
Microinjection In this method, foreign DNA is directly injected into the nucleus of animal cell-or plant cell by using microneedles or micropip’ettes without any use of a vector DNA. It is used to v deliver a transgene or foreign DNA into the egg. Microinjection is especially used for transferring the foreign DNA in animal cells. The transgene is injected into the male pronuclei because it is larger and traceable with a dissecting microscope.
The male pronucleus is then fused with female pronucleus to produce zygote in vitro. In plant cells, the cellulose cell wall is first removed by enzymatic digestion before micro injection. This process is very tedious and involves a lot of precision.

Question 15.
Describe briefly electroporation.
Answer:
Electroporation It is the formation of temporary pores in the plasma membrane of host cells by using lysozyme or calcium chloride under high voltage of electric current. These pores are used for introduction of foreign DNA.
It can be used to transform plant protoplast, i.e. the plant cell with removed cell wall.
Question 16.
What is Polymerase Chain Reaction (PCR)?
Answer:
Polymerase Chain Reaction (PCR)
It is one of the important technique that serves the purpose of amplification of nucleic acid without using a host cell. PCR was discovered by Kary B Mullis in 1983.
Write brief notes on the following
Question 1.
Restriction endonuclease
Answer:
Restriction endonuclease These are specific enzymes, which recognise specific sequences called recognition sequences on DNA and make double-stranded cuts either within the recognition sequence or at a variable distance from the recognition sequence.
They act as scissors and therefore, often are referred to as molecular scissors. The point of cleavage is known as a restriction site. There will be as many restriction sites as the number of recognition sites. The REs that are used in the recombinant DNA technology fall under class-II. These make double-stranded symmetrical cuts within recognition sequences, generating cohesive or blunt ends.
![]()
Question 2.
DNA Ligases
Answer:
DNA Ligases
These enzymes help to join or seal 3-OH end of one nucleic acid fragment with the 5′-P end of another nucleic acid fragment by forming a phosphodiester bond. As they are able to stick DNA fragments together, they are known as molecular glue. The widely used ligase enzyme is T4 DNA ligase. It is purified from E. coli cells that are infected by T4 bacteriophages.
Question 3.
DNA polymerase
Answer:
DNA polymerase enzyme polymerises the DNA synthesis on DNA template or complementary DNA (cDNA). It was discovered by A. Kornberg and co-workers in E. coli in 1956 in the presence of a preformed DNA template, it produces a parallel strand in the presence of ATP.
Question 4.
Southern blotting
Answer:
When the blotting is applied to DNA it is known as Southern blotting (named so after its discover EM Southern in 1975).
Following this, complementary DNA or RNA sequence (probe) labelled with 32 P (radioactive phosphorus) are, hybridised.
The probe binds its complementary target DNA sequence, by forming hydrogen bonds and a duplex is formed. This process is known as molecular hybridisation. The procedure that combines Southern blotting and molecular hybridisation is known as Southern blot hybridisation. After this, the target DNA sequence is identified using autoradiography.
Question 5.
Agarose gel electrophoresis
Answer:
Agarose Gel Electrophoresis:
The molecules of nucleic acids are given uniform negative charge in alkaline buffer solution. These molecules then migrate towards the anode. The smaller molecules migrate faster than larger molecules because the rate of migration is inversely proportional to the molecular weight of separating molecules and directly proportional to the strength of electric field.
Question 6.
Cloning plasmid
Answer:
Plasmids were discovered by William Hayes and Joshua Lederberg (1952). These are extrachromosomal, self-replicating, usually circular, double-stranded DNA molecules, found naturally in many bacteria and also in some yeast. Although plasmids are usually not essential for normal cell growth and division, they often confer some traits on the host organism. For example, resistance to certain antibiotics or toxins that can be a selective advantage under certain conditions.
The plasmid molecules may be present as 1 or 2 copies or in multiple copies (500-700) inside the host organism. These naturally occurring plasmids have been modified to serve as vectors in the laboratory. The most widely used, versatile, easily manipulated vector pBR 322 is an ideal plasmid vector.
Question 7.
Cosmid
Answer:
Cosmid (cos + plasmid) vectors The term cosmid is a combination of two words. COS + MID. COS is taken from COS site of Lambda phage and MID is taken from plasmid DNA. Cosmid was developed for the first time by Collins and Honn (1978). The simplest cosmid vector contains a plasmid’s origin of replication, a selectable marker, suitable restriction enzyme sites and the lambda ‘cos’ site. Cosmids can be used to clone DNA fragments of upto 45 kb length.
![]()
Question 8.
Recombinant DNA
Answer:
Recombinant DNA is the DNA that has formed artificially by genetic engineering. It contain the gene of interest and it becomes the part of host’s genetic makeup where it further replication. The process of constructing recombinant DNA is called Recombinant DNA Technology (RDT).
Question 9.
Polymerase chain reaction
Answer:
A gene can also be cloned or amplified without the assistance of a host cell by a specific reaction, known as Polymerase Chain Reaction (PCR). PCR was discovered by Kary B. Mullis in 1983. The reaction is carried out in a thermocycler having a thermal cycling (heating and cooling) programme.
The target gene is put along with other substrates in the thermocycler. The DNA fragment, doubles at the end of the first cycle and quadruples after the second cycle and so on. In this amplification process, no host cell is used as the supporting system. It is carried out in three steps-denaturation, primer annealing and extension.
Question 10.
Micro injection
Answer:
Microinjection In this method, foreign DNA is directly injected into the nucleus of animal cell-or plant cell by using microneedles or micropip’ettes without any use of a vector DNA. It is used to v deliver a transgene or foreign DNA into the egg. Microinjection is especially used for transferring the foreign DNA in animal cells. The transgene is injected into the male pronuclei because it is larger and traceable with a dissecting microscope.
The male pronucleus is then fused with female pronucleus to produce zygote in vitro. In plant cells, the cellulose cell wall is first removed by enzymatic digestion before micro injection. This process is very tedious and involves a lot of precision.

Long Answer Type Questions
Question 1.
Describe briefly about the making of a recombinant DNA.
Or Describe briefly recombinant DNA technology.
Answer:
Recombinant DNA Technology
Genetic engineering is alternately called as recombinant DNA technology or gene cloning. Paul Berg (1972) was ‘ awarded Nobel Prize in 1980 and is considered as father of genetic engineering. The first recombinant DNA (rDNA) was constructed by Stanley Cohen and Herbert Boyer in 1972. Recombinant DNA technology involves the following steps
1. Isolation of the genetic material (DNA) is carried out as follows
(a) DNA is enclosed within the membranes. To release DNA along with other macromolecules -such as RNA, proteins, polysaccharides and lipids, bacterial cells/plant or animal tissue are treated with enzymes such as lysozyme (bacteria), cellulase (plant cells), chitinase(fungus).
(b) Other molecules can be removed by appropriate treatments and purified DNA ultimately precipitates out as fine threads in the suspension after the addition of chilled ethanol.
2. Cutting of DNA at specific locations is done by using restriction enzymes. The purified DNA is incubated with the specific restriction enzyme at conditions optimum for the enzyme to act.
3. Isolation of desired DNA fragment is carried out using agarose gel electrophoresis.
4. Amplification of gene of interest using Polymerase Chain Reaction (PCR) is a reaction in which multiple copies of specific DNA (gene of interest) sequence are made (amplification) in-vitro.
5. Ligation of DNA fragment into a vector requires a vector DNA and source DNA.
(a) These are cut with endonuclease to obtain sticky ends.
(b) Both are then ligated by mixing vector DNA, gene of interest and enzyme DNA ligase to form recombinant DNA.
6. Insertion of recombinant DNA into the host cell/organism occurs by several methods, before which the recipient cells are made competent to receive the DNA.
(a) If recombinant DNA carrying antibiotic resistance gene (e.g. ampicillin), is transferred into E. coli cells, the host cell is transformed into ampicillin resistant cell.
(b) The ampicillin resistant gene can be called a selectable marker.
(c) When transformed cells are grown on agar plates containing ampicillin, only transformants will grow and others will die.
7. Culturing the host cells The cell containing the foreign gene is cultured on an appropriate medium at optimal conditions. The DNA gets multiplied.
8. Extraction of desired gene product is carried out in the following steps
(a) A protein encoding gene expressed in a heterologous host is called recombinant protein.
(b) Cells having genes of interest can be grown on a small or on a large scale.
