Odisha State Board CHSE Odisha Class 12 Biology Solutions Chapter 7 Molecular Basis of Inheritance Textbook Questions and Answers.
CHSE Odisha 12th Class Biology Chapter 7 Question Answer Molecular Basis of Inheritance
Molecular Basis of Inheritance Class 12 Questions and Answers CHSE Odisha
Very Short Answer Type Questions
Fill in the blanks with correct answers from the choices given in the brackets of each bit
Question 1.
In split genes, the coding sequences are ……….. (introns, operons, exons, cistrons)
Answer:
exons
Question 2.
The smallest part of the gene is called …………… (recon, muton, exon, cistron)
Answer:
cistron
Question 3.
The enzyme referred to as Kornberg enzyme is ……….. (DNA polymerase-I, DNA polymerase-II, RNA polymerase, ligase)
Answer:
DNA polymerase-I
Question 4.
The polymerase that has 5′-3′ exonuclease property is known as ………… (DNA pol-I, DNA pol-II, RNA pol, DNA ligase)
Answer:
DNA pol-I
Question 5.
The termination factor that recognises the termination codon UAG is ………….. (only RF1, only RF2, both RF1 and RF2, neither RF1 and RF2)
Answer:
Only RF
Question 6.
The enzyme that removes formyl group from the first amino acid methionine of a newly synthesised polypeptide is ………… (RF3 translocase, deformylase, exoaminopeptidase)
Answer:
deformylase
Question 7.
The word gene was coined by ……….. (Garrod, Johannsen, Meischer, Griffith)
Answer:
Wilhelm Johannsen
Question 8.
In 1869 …………… discovered DNA.(Garrod, Meischer, GrifFith, Wilkins)
Answer:
Friedrich Meischer
Question 9.
The virulent, pneumococcus possessed a …………. coat for its protection. (protein, lipid, phospholipid, polysaccharide)
Answer:
polysaccharide
Question 10.
Complete sequence of amino acids in ………….. was proposed by Sanger. (insulin, haemoglobin, kinetin, polymerase)
Answer:
insulin
Question 11.
RNAs lack ………. as nitrogenous base. (adenine, guanine, cytosine, thymine)
Answer:
thymine
Question 12.
One complet turn of B-DNA contains ………….. number of nitrogenous bases. (10,11,9,12)
Answer:
10 .
Question 13.
The most stable form of RNA is …………. RNA. (messenger, transfer, ribosomal, small nuclear)
Answer:
ribosomal
Question 14.
When a codon codes or more than one amino acid, it is called ………….. code.
(commaless, degenerate, nonsense, universal)
Answer:
degenerate
Question 15.
The start codon is ………… (UAA, UGA, AUG, UGA)
Answer:
AUG
Express in one or two word(s)
Question 1.
If in a double-stranded DNA there is 25% of thymine, then calculate the per cent of guanine.
Answer:
25%
Question 2.
What is the complementary base of adenine in RNA?
Answer:
Uracil
Question 3.
In a double helix if one stand is on 5′ → 3′, what will be arrangement of other strand?
Answer:
3′ → 5′
Question 4.
What are the basic proteins called in eukaryotic DNA?
Answer:
Histones
Question 5.
What is called to amino acids with more than one codon?
Answer:
Degenerate
Question 6.
What type of genes do express continuously ?
Answer:
Housekeeping genes
Question 7.
What type of RNAs do carry amino acids to the site of protein synthesis ?
Answer:
tRNA
Correct the sentences in each bit without changing the underlined words
Question 1.
Watson and Griffith proposed double helical structure of DNA.
Answer:
Watson and Crick proposed the double helical structure of DNA.
Question 2.
A nucleoprotein is building block of all nucleic acids.
Answer:
Nucleotides are the building blocks of all nucleic acids.
Question 3.
The strand of the DNA double helix represent nucleotide phosphate backbone and are antiparallel.
Answer:
The DNA double helix consist of a polynucleotide chain with a backbone formed of sugar and phosphate groups. These chains are antiparellel to each other.
Question 4.
The helical turns are right handed is Z DNA.
Answer:
The helical turns are left-handed in Z-DNA.
Question 5.
Avery, McCarty and MacLeod experimentally proved that the transforming principle is a protein.
Answer:
Avery, McCarty and MacLeod experimentally proved that transforming principle is DNA.
Question 6.
Meischer proposed the transforming principle.
Answer:
Frederick Griffith proposed the transforming principle.
Question 7.
The enzyme ligase is responsible for transcription.
Answer:
The enzyme DNA dependent RNA polymerase is responsible for transcription.
Question 8.
The operator is under the control of a repressor molecule synthesised by structural gene which is not a part of operon.
Answer:
The operator is under the control of a repressor molecule synthesised by regulator gene (i-gene) which is a part of operon.
Question 9.
The example of regulatory gene is genes of respiratory enzymes.
Answer:
The example of constitutive gene is genes of respiratory enzymes.
Question 10.
P-site in prokaryotes only accepts tRNAmet.
Answer:
P-site in prokaryotes only accepts tRNA fmet.
Question 11.
The coding or translatable sequences are introns.
Answer:
The intervening or non-coding sequences are introns.
Question 12.
The structural gens transcribe tRNA and rRNA.
Answer:
The structural gene do not transcribe tRNA and rRNA.
Question 13.
A primer is a small DNA or RNA strand hydrogen bonded to a template.
Answer:
A primer is a short strand of RNA or DNA that serves as a starting point for DNA synthesis.
Question 14.
In DNA replication, as per semiconservative model, two new strands synthesised, form new DNA molecules.
Answer:
In DNA replication as per semiconservative model, two parental strands would separate and act as a template for the synthesis of new complementary strands. Hence, a new strand consist of one parental strand and other new replicated strand.
Fill in the blanks
Question 15.
The enzyme …………. hydrolyses DNA molecules.
Answer:
exonucleases
Question 16.
Clover leaf model of fRNA was proposed by ………………
Answer:
Holey
Question 17.
The segment of DNA that expresses specific character is called …………..
Answer:
gene
Question 18.
The enzyme ………… helps to join nucleotides.
Answer:
ligase
Question 19.
The DNA strand which takes part in transcription is called ………………
Answer:
leading strand
Question 20.
UAG is …………. codon.
Answer:
stop
Question 21.
The gene which becomes active due to the presence of specific substance is called ………….. gene.
Answer:
inducible
Question 22.
To identify criminals DNA ………… is done.
Answer:
fingerprinting
Short Answer Type Questions
Write notes on the following with atleast 2 valid points
Question 1.
Inducible operon
Answer:
Inducible operon An inducible operon is an operon, in which the presence of a key metabolic substance induces transcription of the structural genes. One example of an inducible operon is lac operon and the inducer of this operon is lactose.
Question 2.
Repressible operon
Answer:
Repressible operon A repressible operon is an operon which always transcribes structural genes unless a repressor is present. One example of a repressible operon is trp operon.
Question 3.
Housekeeping genes
Answer:
House keeping genes are involved in basic cell maintenance and therefore are expected to maivlain constant constant expression levels in all cells and conditions. They are also called constituline gene
Question 4.
Adaptor molecules
Answer:
Adapter molecules tRNA acts as adapter molecules in translation. It serves as the physical link between the mRNA and the amino acid sequence of proteins. It carries an amino acid to the ribosome as directed by a three nucleotide sequence in a mRNA.
Question 5.
Split genes
Answer:
Split genes It is an interrupted gene that contains sections of DNA called exons which are expressed as RNA and proteins, interrupted by sections of DNA called introns, which are not expressed.
Question 6.
RNA splicing
Answer:
RNA splicing Splicing is the editing of the nascent precursor of mRNA transcript into a mature mRNA. After splicing, introns are removed and exons are joined together. The process of splicing involves two successive transesterification reaction. The RNA splicing is carried out with the help of a large complex called spliceosome.
Question 7.
Termination of translation
Answer:
Termination:
It is accomplished by the presence of any of the three termination codon on mRNA. Which are recognised by the release (termination) factor, RF1, RF2 and RF3.
RF1 and RF2 They resemble the structure of tRNA. They compete with tRNA to bind the termination codon at A-site of ribosome. This is called molecular mimicry. RF1 recognise UAG and RF2 recognise UGA. UAA is recognised by both of them. RF3 helps to split the peptidyl tRNA body by using GTP.
Note In eukaryotes, only one release factor is known. It iseRF1.
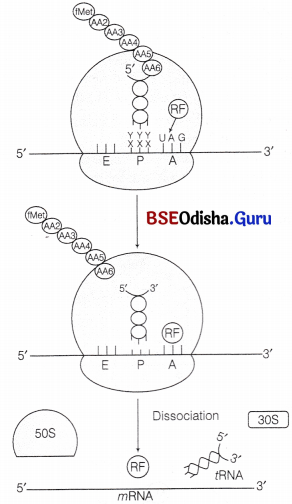
Question 8.
Okazaki fragments
Answer:
Okazaki fragments During DNA replication, the new strand elongation occurs in opposite directions, as the two strands of the DNA are antiparallel.
On the leading strands, a continuous synthesis of new strand occurs while on the other strand (5′ → 3′ strand) discontinuous synthesis occurs. These discontinuous pieces of new DNA strands are called Okazaki fragments which join by DNA ligase to form the continuous strand
Question 9.
Central dogma
Answer:
Central Dogma:
It was proposed by Francis Crick (in 1958). According to the central dogma in molecular biology, the flow of genetic information is unidirectional, i.e.
DNA → RNA → Protein.

Central dogma
But later in 1970, HM Temin reported that the flow of information can be in reverse direction also, i.e. from RNA to DNA in some viruses (e.g. HIV) which is called as reverse transcription.
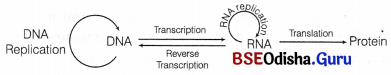
Modified central dogma
Note: Eukaryotic gene expressions were obtained from yeast,
Arabidopsia thaliana with developed techniques.
Differentiate with atleast 2 valid points
Question 1.
Genes and Chromosomes.
Answer:
Differences between genes and chromosomes are as follows
| Genes | Chromosomes |
| Gene is a segment of DNA on the chromosome that codes for a functional protein and RNAs like tRNA, rRNA or ribozymes. | Chromosome is the structure formed by the condensation of chromatin during cell division. |
| Genes basically refers to the DNA fragment that directs the synthesis of a protein. | Chromosome consists of long DNA strand wrapped around histone proteins. |
| Gene contain coding sequence called exons and non-coding sequence called introns on the chromosome that directs synthesis of a protein. | Chromosome is a long DNA strand containing bdth coding (genes) and non-coding DNA (junk DNA or spacer DNA) between genes. |
Question 2.
DNA and RNA.
Answer:
Differences between DNA and RNA are as follows
| DNA | RNA |
| It is mainly confined to the nucleus, but also occurs in mitochondria and chloroplasts in small amount. | It mainly occurs in the cytoplasm. A small quantity is found in the nucleus. |
| It contains deoxyribose. | It contains ribose sugar. |
| It pyrimidines are cytosine and thymine. | Its pyrimidines are cytosine and uracil. |
| It consists of two polynucleotide chains held together by hydrogen bonds and coiled into a double helix. Some viruses (ϕ x 174) have single- stranded DNA. | It consists of a single polynucleotide chain. It may fold on itself and get hydrogen bonded and coiled into a pseudohelix. Some viruses (reovirus) have double-stranded RNA. |
Question 3.
Purines and Pyrimidines.
Answer:
Differences between purines and pyrimidines are as follows
| Purines | Pyrimidines |
| It contains two carbon-nitrogen rings and four nitrogen atoms. | It contains one carbon nitrogen ring and two nitrogen atoms. |
| Purines are adenine and guanine | Pyrimidines are cytosine, thymine and uracil. |
| 120.11 g mol molar mass | 80.088 g mol<sup>-1</sup> molar mass. |
Question 4.
Exons and Introns.
Answer:
Differences between exons and introns are as follows
| Exons | Introns |
| These are the nucleotide sequence of genes that are expressed and those are found at either sides of an intron. | These are sequences of nucleotides present in the genes between exons. |
| These are nucleotide sequences which code for proteins. | These are nucleotide sequences do not code for proteins. |
| These are translated regions on mRNA. | These are untranslated regions on mRNA. |
| After RNA splicing, only exons are present on the mature mRNA. | These are removed from mRNA during RNA splicing. |
Question 5.
B-DNA and Z-DNA.
Answer:
Differences between B-DNA and Z-DNA are as follows
| B-DNA | Z-DNA |
| It is with a right-handed type of helix. | It is with a left-handed type of helix. |
| Its helical diameter is 2.37 nm. | Its helical diameter is 1.84 nm. |
| It has 0.34 nm rise per base pair. | It has 0.37 nm rise per base pair. |
| It has 3.4 nm distance per complete turn. | It has 4.5 nm distance per complete turn. |
| Number of base pair per complete turn are 10. | Number of base pair per complete turn are 12. |
Question 6.
Answer:
Replication and Transcription.
Answer:
Differences between replication and transcription are as follows
| Replication | Transcription |
| It occurs in the S-phase of cell cycle. | It occurs in the G<sub>1</sub> and G<sub>2</sub> phases of cell cycle. |
| It is catalysed by DNA polymerase enzyme. | It is catalysed by RNA polymerase enzyme. |
| Deoxyribonucleoside triphosphate (dATP, dGTP, dTTP) serve as raw materials. | Ribonucleoside triphosphate (ATP, UTP, GTP, CTP) serve as raw materials. |
| Replication occurs along the strands of DNA. | IT takes place along one strand of DNA. |
| It involves unwinding and splitting of the entire DNA molecule. | It involves unwinding and splitting of only those genes which are to be transcribed. |
| It involves copying of the entire genome. | It involves copying of certain individual genes only. |
| Two double-stranded DNA molecules are formed from one DNA molecule. | A single one-stranded RNA molecule is formed from a segment of one strand. |
| Serve to conserve the genome for the next generation of cells and individuals. | Serve to form DNA copies of individual genes for immediate use in protein synthesis. |
| It require RNA primer to start replication. | No primer is required to start. |
| It produces normal DNA molecules that do not need | It produces primary RNA transcript molecules which need processing to acquire final form and size. |
Question 7.
Transcription and Translation.
Answer:
Differences between transcription and translation are as follows
| Transcription | Translation |
| It is the formation of RNA from DNA. | It is the synthesis of polypeptide over ribosome. |
| The template is antisense strand of DNA. | The template is mRNA. |
| It occurs inside the nucleus in eukaryotes and cytoplasm in prokaryotes. | It occurs in cytoplasm. |
| The raw materials are four types of ribonucleoside triphosphates – ATP, GTP, CTP and UTP. | The raw materials are 20 types of amino acids. |
| It forms three types of RNAs, i.e. rRNA, tRNA and mRNA | All the three types of RNAs take part in translation. |
| Transcription requires RNA polymerases and some transcription factors. | Translation requires initiation, elongation and releasing factors. |
| Polymerase moves over the template DNA. | Ribosome moves over mRNA. |
| An adapter molecule is not required. | Adaptor molecules bring amino acids over the template. |
| Product often requires splicing. | Splicing is absent. |
Question 8.
Housekeeping gene and Inducible gene.
Answer:
Differences between housekeeping gene and inducible gene are as follows
| Housekeeping gene | Inducible gene |
| These genes are constantly expressing themselves in a cell because their products are required for the normal cellular activities, e.g. genes for glycolysis, ATPase. | These genes are switched on in response to the presence of a chemical substance or inducer which is required for the functioning of the product of gene activity, e.g, nitrate for nitrate reductase. |
Question 9.
Degenerate codon and Nonsense codon.
Answer:
Differences between degenerate codon and nonsense codon are as follows
| Degenerate codon | Nonsense codon |
| Genetic code is degenerate for a particular amino acid, that is more than one codon can code for a single amino acid. | A codon for which no normal tRNA molecule exists does not code for any amino acid. |
| These codon causes translation. | The presence of nonsense codon causes termination of translation ending polypeptide synthesis chain. |
| These are many e.g. phenylalanine has two Codon is UAA and UUC. | These are three nonsense codons and are called amber(UAG), ochre(UAA) and opal(UGA). |
Long Answer Type Questions
Question 1.
Give the structure of DNA. Add a note on different forms of DNA.
Answer:
Primary Structure of DNA:
Two nucleotides when linked through a 3′-5′ phosphodiester linkage, form a dinucleotide. The phosphodiester linkage is formed when each phosphate group esterifies to the 3′ hydroxy group of a pentose and to the 5′ hydroxyl group of the next pentose.
In a similar fashion, more nucleotides may join to form a polynucleotide chain (fig. structure of DNA). The polymer chain thus, formed has
(i) One end with a free phosphate moiety at 5′ end of deoxyribose sugar. This is marked as 5′ end of polynucleotide chain.
(ii) The other end with a free hydroxyl 3′-OH group marked as 3′ end of the polynucleotide chain.
Thus, the sugar and phosphates form the backbone in a polymer chain and the nitrogenous bases linked to sugar moiety project from this backbone. In RNA, there is an additional -OH group at 2′ position in the ribose of every nucleotide residue.
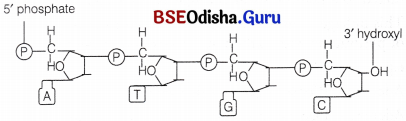
Secondary Structure of DNA:
Watson and Crick proposed the secondary structure in the form of the famous double helix model in 1953 on the basis of following observations
1. Erwin Chargafif (in 1950) formulated important generalisation on the base and other contents of DNA, called as ChargafFs rule. It states that for a double-stranded DNA, the ratios between adenine (A) and thymine (T) and guanine (G) and cytosine (C) are constant and equal to one.
i.e. \(\frac{A + T}{G + C}\) = 1
2. X-ray diffraction studies by Wilkins in 1952, suggested a helicoidal configuration of DNA.
One of the important features of this model was the complementary base pairing. It means if the sequence of bases in one strand is known, the sequence in other strand can be easily predicted. Also, if each strand from a DNA acts as a template for synthesis of a new strand, the daughter DNA thus produced would be identical to the parental DNA molecule.
Watson and Crick Model of DNA:
Watson and Crick worked out the first correct double helix model of DNA, which explained most of its properties.
The salient features of double helix structure of DNA are as follows
(i) DNA is made up of two polynucleotide chains. The backbone is constituted by sugar phosphate, while the nitrogenous bases project inwards.
(ii) The two chains have anti-parallel polarity, i.e. when one chain has 3′ → 5′ polarity, the other has 5′ → 3′ polarity. Hence, orientation of deoxyribose sugar is opposite in both the strands.
(iii) The two strands are complementary to each other, i.e. purine base of one strand has pyrimidine counterpart on other strand. The complementary bases in two strands are paired through hydrogen bonds (H-bonds) to form base pairs.
(a) Adenine is bonded with thymine of the opposite strand with the help of two hydrogen bonds.
(b) Guanine is bonded with cytosine of the opposite strand with the help of three hydrogen bonds. So, a purine bonds with a pyrimidine always. Thus, maintaining a uniform distance between the two strands of the helix.
(iv) The two polypeptide chains are coiled in a right-handed fashion. Pitch of the helix, i.e. length of DNA in one complete turn = 3.4 nm or
3.4 × 10-9 or 34 Å.
Number of base pairs in each turn = 10. Distance between a base pair in a helix = 0.34 nm. The diameter of DNA molecule is 20 Å (2nm).
(v) Percentage calculation of bases is done by
A + T = 100 – (G + C).
(vi) The plane of one base pair stacks over the other in double helix. This provides the stability to the helical structure, in addition to H-bond.
The length of DNA in E. coli is 1.36 mm, while in humans it is 2.2 m.
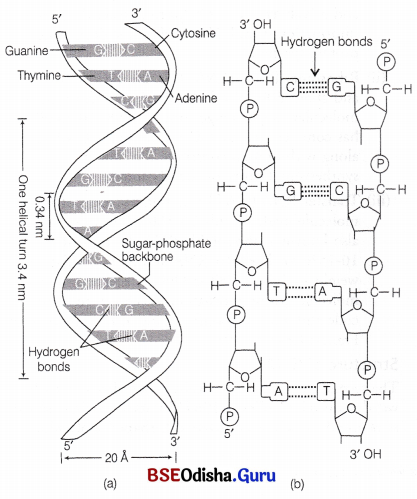
Structure of DNA : (a) Watson and Crick model of double helix, (b) Double-stranded polynucleotide chain sequence showing hydrogen bonds
Structural Forms of the Double Helix:
Double helical DNA exists in three structural forms namely the A-form DNA, the B-form DNA (described by Watson and Crick) and the Z-form DNA. The transition among these three forms plays an important role in the regulation of gene expression.
(i) B-DNA It is right-handed helix, contains 10 residues per 360° turn.
- The planes of bases are perpendicular to the helix axis.
- It is primarily found in chromosomal DNA.
(ii) A-DNA
- It is formed by the moderate dehydration of B-DNA.
- It is right-handed helix, contains 11 base pairs (residues) per 360° turn.
- The planes of bases are tilted 20° away from the perpendicular to helical axis.
- It is mainly found in DNA-RNA hybrid or RNA-RNA double-stranded regions.
(iii) Z-DNA It has zig-zag backbone and hence has the name.
- It is a left-handed helix, twelve base pairs are present per turn.
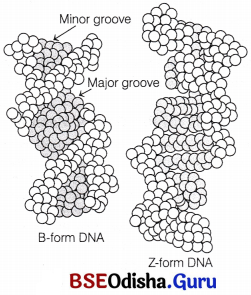
- The Z-DNA stretches occurring naturally in DNA have a sequence of alternating purines and pyrimidines (i.e. poly GC regions).
- Besides these three major forms, the other two right-handed forms are
- C-DNA with nine base pairs per turn.
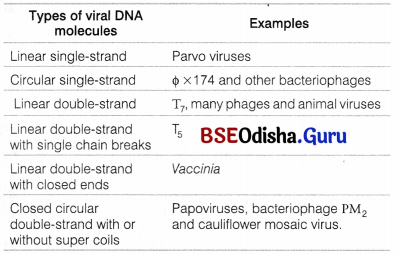
- D-DNA with eight base pairs per turn. There are many forms of DNA molecules in viruses.
Various Forms of DNA Molecules Found in a Variety of Viruses
Question 2.
Describe the semiconservative model of DNA replication.
Answer:
DNA Replication
In addition to the double helical structure of DNA, Watson and Crick also proposed a scheme for DNA replication. According to this model, the two strands of double helix separate and act as a template for the synthesis of new complementary strands in which the base sequence of one strand determines the sequence on the other strand.
This is called base complementarity and it ensures the accurate replication of DNA. After the completion of replication, each DNA molecule have one parental and one newly synthesised strand.
This scheme for DNA replication was termed as semiconservative DNA replication.
DNA Replication is Semiconservative:
Matthew, Meselson and Franklin Stahl in 1958 conducted an experiment in California Institute of technology with Escherichia coli to prove that DNA replicates semiconservatively as follows
1. They grew many generations of E.coli in a medium containing 15NH5Cl (15N is the heavy isotope of nitrogen) as the only source of nitrogen.
The result was that 15N got incorporated into the newly synthesised DNA after several generations. By centrifugation in a cesium chloride (CsCl) density gradient, this heavy DNA molecule could be distinguished from the normal DNA.
2. The bacterial culture was then washed to make the medium free and the cells were then transferred into a medium containing normal 14NH4Cl.
3. The samples were separated independently on CsCl gradient to measure the densities of DNA.
4. At definite time intervals, as the cells multiplied, samples were taken and the DNA which remained as double-stranded helices was extracted.
5. The DNA obtained from the culture, one generation after the transfer from 15N to 14N medium (i.e. after 20 minutes because E.coli divides in 20 minutes) had a hybrid or intermediate density.
This hybrid density was the result of replication in which DNA double helix had separated and the old strand (N15) had synthesised a new strand (N14).
6. The DNA obtained from the culture after another generation (II generation) was composed of equal amounts of hybrid DNA containing (N15 molecule) and ‘light’ DNA (containing 4 N molecule).
7. With the preeceding growth generations in normal medium, more and more light DNA was present. Hence, the semiconservative mode of DNA replication was confirmed.
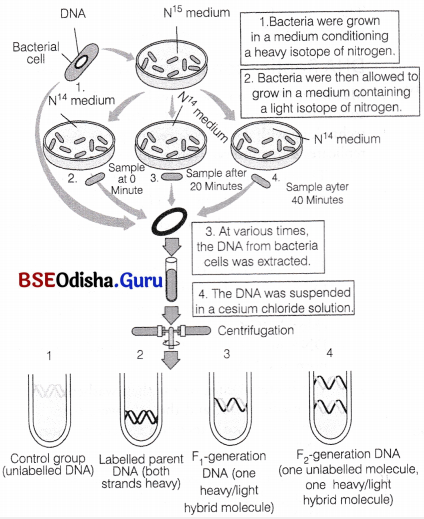
Meselson-Stahl experiment to demonstrate semiconservative replication
were conducted by Taylor and colleagues in 1958, involving use of radioactive thymidine, i.e. bromodeoxy-uridine. The replicated chromosomes were then, stained with fluorescent dye and Giemsa. The old and new strand were stained differently which confirm that the DNA in chromosomes also replicates semiconservatively.
Question 3.
Give evidences of DNA as genetic material.
Answer:
DNA as Genetic Material:
The discovery of nuclein by Meischer and the proposition of principal of inheritance by Mendel were almost at the same time, i.e. 1869 and 1866, respectively. But the fact that DNA acts as a genetic material took a long time to be discovered and proven.
By 1926, the quest to determine the mechanism for genetic inheritance had reached the molecular level and gradually the question, what molecule acts as genetic material got answered.
Transforming Principle:
Frederick Griffith in 1928, carried out a series of experiments with Diplococcus pneumoniae (a bacterium that causes pneumonia). He observed that when these bacteria were grown on a culture plate, some of them produced smooth, shiny colonies (S-type), whereas the others produced rough colonies (R-type).
This difference in appearance of colonies (smooth/rough) is due to the presence of mucous (polysaccharide) coat on S-strains (virulent/pathogenic) but not on R-strains (avirulent/non-pathogenic).
Experiment:
- He first infected two separate groups of mice. The mice that were infected with the S-strain (S-III) died from pneumonia as S-strains are the virulent strains causing pneumonia.
- The mice that were infected with the R-strain (R-II) did not develop pneumonia and they lived.
- In the next set of experiments, Griffith killed the bacteria by heating them. The mice that were injected with heat-killed S-strain bacteria did not die and lived.
- Whereas, on injecting a mixture of heat-killed S-strain and live R-strain bacteria, the mice died. Moreover, living S-bacteria were recovered from the dead mice.
These steps are summarised below

From all these observations Griffith concluded that the live R-strain bacteria, had been transformed by the heat-killed S-strain bacteria, i.e. some ‘transforming principle’ had transferred from the heat-killed S-strain, which helped the R-strain bacteria to synthesise a smooth polysaccharide coat and thus, become virulent.
This must be due to the transfer of the genetic material. However, he was not able to define the biochemical nature of genetic material from his experiments.
Biochemical Characterisation of Transforming Principle:
Oswald Avery, Colin MacLeod and Maclyn McCarty (1933-44) worked in Rockfellar Institute, New Xork, USA to determine the biochemical nature of ‘transforming principle’ in Griffith’s experiment in an in vitro system. Prior to this experiment, the genetic material was thought to be protein.
During this experiment, purified biochemicals (i.e. proteins, DNA, RNA, etc.) from the heat-killed S-III cells were taken, to observe which biochemicals could . transform live R-cells into S-cells.
They discovered that DNA alone from heat-killed S-type bacteria caused the transformation of non-virulent R-type bacteria into S-type virulent bacteria.
They also discovered that protein digesting enzymes (proteases) and RNA digesting enzymes (ribonuclease) did not inhibit this transformation. This proved that the ‘transforming substance’ was neither protein nor RNA.
DNA-digesting enzyme (deoxyribonuclease) caused inhibition of transformation, which suggests that the DNA caused the transformation. This provided the first evidence for DNA as transforming principle or the genetic material.
The steps of this experiment are summarised below:
- R-II + DNA extract of S-III + no enzyme = R-II colonies + S-III colonies
- R-II + DNA extract of S-III + Ribonuclease = R-II colonies + S-III colonies
- R-II + DNA extract of S-III + Protease = R-II colonies + S-III colonies
- R-II + DNA extract of S-III + Deoxyribonuclease = Only R-II colonies
Question 4.
Explain the mechanism of translation in prokaryotes.
Answer:
Mechanism of Translation:
The main steps in translation include
(i) Binding of iwRNA to ribosome
(ii) Activation of amino acids (aminoacylation of rRNA).
(iii) Transfer of activated amino acids to rRNA.
(iv) Initiation of polypeptide chain synthesis.
(v) Elongation of polypeptide chain.
(vi) Termination of polypeptide chain formation.
(i) Binding of mRNA to Ribosome
Ribosomes occur in the cytoplasm as separate smaller and larger units.
In prokaryotes, IF-3 binds to 30 S subunit to prevent association of subunits. The incoming tRNA containing specific amino acid binds to the A-site. The prokaryotic mRNA has a leader sequence called shine-Delgarno sequence that is present just prior to initiation codon-AUG.
It is homologous to 3′ end of 16S rRNA (ASD region) in 30S subunit. This complementarity ensures correct binding of 30S subunit on mRNA. The peptidyl mRNA containing elongating polypeptide then binds to P-site.
The bacterial ribosome contains another site, the E-site or Exit-site to which the discharged tRNA or tRNA whose peptidyl has already been transferred binds before its release from ribosome.
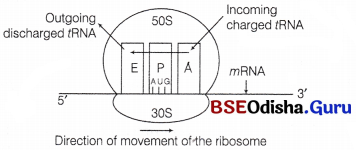
A bacterial ribosome with three sites; E, exit site, P, peptidyl site and A, aminoacyl site
(ii) Aminoacylation
Aminoacylation of tRNA The activation of amino acids takes place through their carboxyl groups. The amino acids are activated in the presence of ATP and linked to their cognate tRNA. In the presence of ATP, amino acids become activated by binding with aminoacyl tRNA synthetase enzyme.

The amino acid AMP-enzyme or AA-AMP enzyme complex is called an activated amino acid.
In the second step, this complex associated with the 3′-OH end of tRNA. AMP gets hydrolysed to form an ester bond between amino acid and tRNA and the enzyme is released.
AA – AMP – enzymes + tRNA ——– AA – tRNA + AMP + enzyme.
(iii) Transfer of Activated Amino Acids to tRNA:
The amino acids are attached to the tRNA by high energy bonds. These bonds are formed between the carboxyl group of amino acid and 3′ hydroxy terminal of ribose of terminal adenosine of CCA and of tRNA.
The complete reaction is carried out by enzyme synthetase which has two active sites, i.e. one for rRNA and another for specific amino acid molecule.
(iv) Initiation of Polypeptide Chain Synthesis:
The protein synthesis begins from the amino terminal end of the polypeptide, proceeds by the addition of amino acids through peptide bond formation and ends at the carboxyl terminal end. In prokaryotes, the initiation amino acid is formylated methionine while in eukaryotes it is methionine.
Initiation in Prokaryotes
In prokaryotes, two types of tRNA are present for methionine
- tRNAfmet for initiation carrying formyl methionine and
- tRNAmet for carrying normal methionine to growing polypeptide.
The initiation of polypeptide synthesis requires the following components
mRNA, 30S subunit of ribosome, formylmethionyl-tRNA (fmet – tRNAfmet), initiation factors IF-1, IF-2 and IF-3, GTP, 50S ribosomal subunit and Mg+2.
The sequence of events occurring during initiation process are
1. The smaller 30S subunit of ribosome binds to the transcription factor IF-3. It prevents the premature association of two ribosomal subunits.
2. Interaction of SD region of mRNA and ASD region of ribosome helps the mRNA to bind to 30S subunit. It also helps AUG to correctly positioned at the P-site of the ribosome.
3. The fMet-tRNAfmet (the specific tRNA aminoacylated to formyl methionine) binds to the AUG codon at the P-site. The tRNAfmet is the only tRNA that binds to its codon present on the P-site. All other tRNA along with their respective amino acids bind to their codon present at the A-site. Therefore, AUG codon present as initiation codon codes for formylmethionine. When it is present at other position it codes for normal methionine.
4. The initiation factor IF-1, binds to the A-site. It prevents the binding of any other aminoacyl tRNA to the codon at the A-site during initiation.
5. The GTP bound IF-2 (GTP-IF-2) and the initiating f Met-tRNAfmet attaches to the complex of 30S subunit-IF3-IF1-mRNA.
6. 50S subunit then attaches the complex formed in the previous step. The GTP bound to IF-2 is hydrolysed to GDP and Pi. After this step, all the three initiation factors leave ribosome. This complex of 70S ribosome, mRNA and f Met-tRNA fmet bound to initiation codon at P site is known as initiation complex.
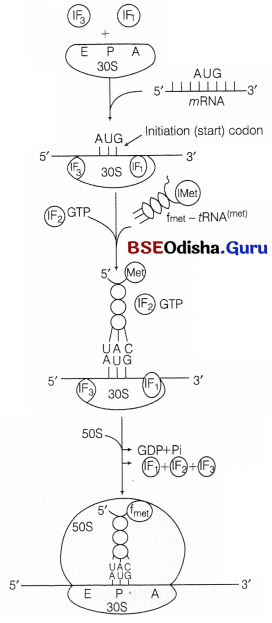
Stepwise formation of initiation complex in prokaryote
(v) Elongation of Polypeptide Chain:
In this step, another charged aminoacyl tRNA complex binds to the A-site of the ribosome, following the hydrolysis of GTP to GDP and Pi. A peptide bond forms between carboxyl group (—COOH) of amino acid at P-site and amino group (—NH3) of amino acid at A-site by the enzyme peptidyl transferase.
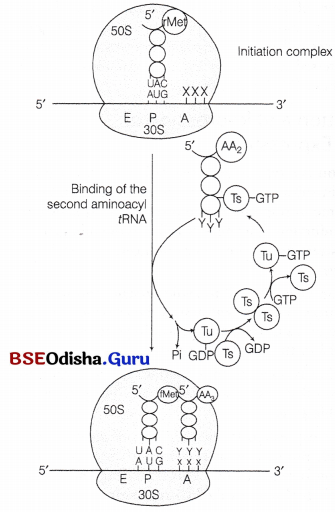
Binding of the second aminoacyl tRNA to the A site of ribosome
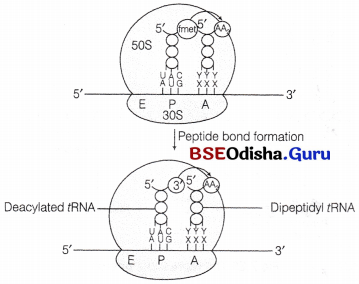
Formation of a peptide bond
(vi) Translocation of Polypeptide:
The peptidyl fRNA bounded to A-site comes to the P-site of ribosome.
The empty tRNA comes to E-site and a new codon occupies the A-site for next aminoacyl tRNA.
- This is achieved by the movement or translocation of ribosome by a codon in 5′ to 3′ direction of mRNA in the presence of EF-G (translocase) and GTP.
- tRNA interact with E-site on 50S subunit through it CCA sequence at 3′ end.
The tRNA molecule is then, transferred from A site to P-site and from P-site to E-site by the movement of two subunits of ribosomes.
Finally, the deacylated tRNA is released to cytosol from E-site.
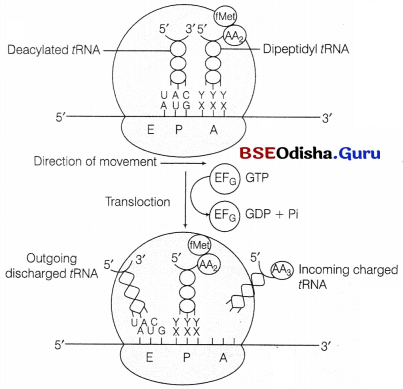
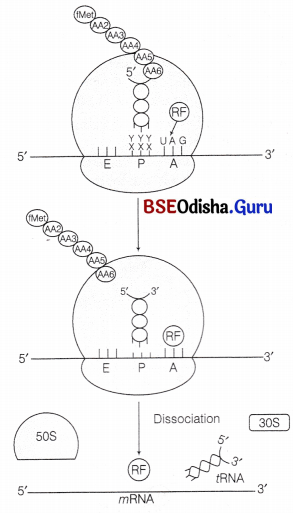
(vii) Termination:
It is accomplished by the presence of any of the three termination codon on mRNA. Which are recognised by the release (termination) factor, RF1, RF2 and RF3.
RF1 and RF2 They resemble the structure of tRNA. They compete with tRNA to bind the termination codon at A-site of ribosome. This is called molecular mimicry. RF1 recognise UAG and RF2 recognise UGA. UAA is recognised by both of them. RF3 helps to split the peptidyl tRNA body by using GTP.
Question 5.
Describe transcription in prokaryotes.
Answer:
Transcription in Prokaryotes:
All three RNAs are needed for synthesis of a protein in a cell. DNA dependent RNA polymerase is the single enzyme that catalyses the transcription of all types of bacterial RNA. But for the expression of different genes, different sigma factors may associate with same core enzymes.
In E.coli, σ70 is used in normal condition σ32 / σH under heat shock, σ54 /σN under nitrogen starvation and σ28 for chemotaxis.
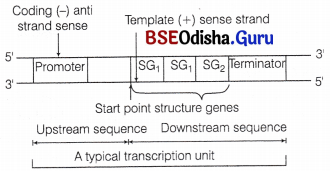
A typical bacterial transcription unit
The transcription process in prokaryotes occurs in following steps
I. Initiation
- The holoenzyme binds to the promoter region of transcription unit.
- The sigma polypeptide binds loosely to the promoter sequences so as to form a loose, closed, binary complex.
- It is followed by the formation of a transcription eye or bubble due to the denaturation of adjacent sequence of DNA, lying next to the complex.
- The transcription bubble along with the bounded holoenzyme is called open binary complex.
- In 90% of cases, the start point of transcription is a purine.
- At the elongation site of enzyme, two nucleotides complementary to the first two nucleotides of template strand binds.
- A phosphodiester bond is formed between these two ribonucleotides.
- At this stage, the complex is called ternary complex that consists of partly denatured DNA bounded with holoenzyme having a di-ribonucleotide.
- The same process continues till a RNA chain of about nine nucleotides is synthesised. The holoenzyme does not move throughout this process.
- After the completion of initiation process, sigma factor dissociates from RNA polymerase. This facilitates the promoter clearance so that a new holoenzyme can bind to promoter for second round of transcription.
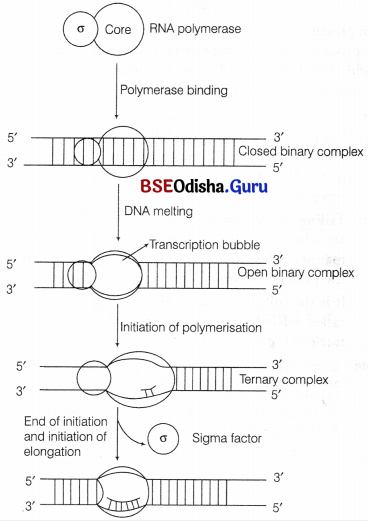
Binding of RNA polymerase and initiation of RNA synthesis
II. Elongation
- The RNA chain grows in 5′-3′ direction by the addition of ribonucleotides to the 3′-end of RNA.
- The transcription bubble moves in 3′ → 5′ direction of template strand.
- The movement of holoenzyme along the bubble unwinds (denature) the DNA in growing point and rewinds at the opposite end.
- In each elongation cycle, the growing site (leading product) of enzyme gets filled with 10 newly added nucleotides. The opposite site (lagging product) contains the previous segment of RNA.
- About 40 nucleotides are added per 37°C.
- In some phages, e.g. T3 and T4, RNA pol synthesise RNA at much rapid rate of about 200 nucleotides per 37° C.
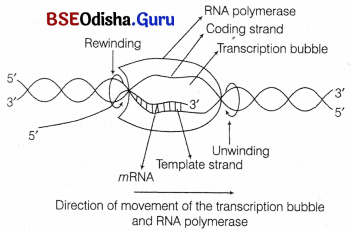
Elongation of RNA chain by the movement of RNA polymerase and transcription bubble in 5′ to 3′ direction
III. Termination
It is achieved by certain termination signals on DNA called terminators. In E.coli, terminators are of two types
(a) Intrinsic Terminators
- These are rho independent protein factors in which the RNA at 3′-end contains a long stretch of U residues.
- These residues are hydrogen bonded to the long stretch of A residues of the temple. In the stem of the RNA, a stretch of G-C rich segment is present which results in a hair-pin loop formation in the RNA stem.
- Due to the weak association between A-U base pairs, the long stretch of termination sequence break and the RNA is released.
- It occurs due to the formation of hair-pin loop in the stem of RNA before the termination signal slows down transcription and as a result dA-rU bonds break at any one point so as to release RNA from RNA-DNA hybrid.
(b) Extrinsic Terminator
- These are rho dependent protein factors and are extensively used in E. coli.
This protein is active as an hexamer (having six identical subunits). Its molecular weight is about 46,000 and it also has ATP hydrolysing activity. - To terminate the process of transcription, rho factor binds to the 5′-end of nascent mRNA and moves along the length of mRNA until it reaches the termination point. Due to this, the transcription process slows down, rho breakdown ATP and utilises the energy to denature the RNA-DNA hybrid. Hence, the RNA is released from the bubble.
In prokaryotes (bacteria), mRNA does not require any processing to become active and both transcription and translation take place in same compartment (as there is no separation of nucleus and cytosol in bacteria). Therefore, translation can start much before the mRNA is fully transcribed, i.e. transcription and translation can be coupled.
Question 6.
Give an account of the operon model.
Answer:
The Operon Model:
F Jacob and J Monod gave the operon concept and were the first ones to describe a transcriptionally regulated system. An operon is a unit of prokaryotic gene expression which includes sequentially regulated (structural) genes and control elements recognised by the regulatory gene product. The various components of an operon are
- Structural genes These are the regions of DNA which transcribe wRNA for polypeptide synthesis.
- Promoter gene This is the sequence of DNA where RNA polymerase binds and initiates transcription.
- Operator This is the sequence of DNA found adjacent to promoter where specific repressor protein binds. It is under the control of a repressor.
- Regulator It is the gene which codes for the repressor protein binding to the operator and suppresses its activity, so that transcription does not occur. It is also represented as ‘i’ gene. It is synthesised by a regulator gene which is not a part a operon.
- Inducer Its main role is to prevent the repressor from binding to the operator. This makes the process of transcription to switch on. An inducer can be any metabolite, hormone, etc.
Loc Operon in E. coli:
The lac operon found in E.coli is an inducible system. It is responsible for the synthesis of enzymes found in lactose (the milk sugar). It has an operator sequence of 26 base pairs and three structural genes. Its first structural gene (SG) is lac Z which is of 3063 base pairs. It is responsible for the synthesis of the emzyme ß-galactosidase.
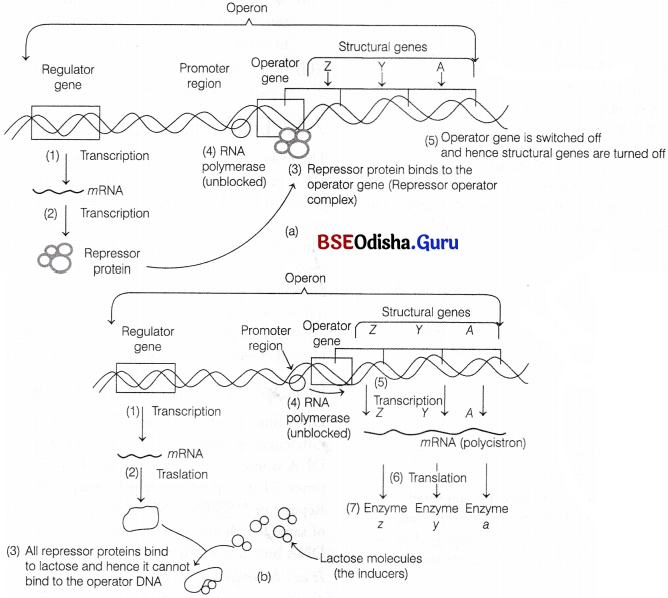
The operator is a part of lac Z. The other two genes are lac y (helps in the synthesis of ß-galactoside permease) and lac a (synthesise fbgalactoside transacetylase). ß-galactoside permease is a transmembrane protein that pumps galactose into the cell, ß-galactosidase helps to breakdown lactose to galactose and glucose.
When lactose is available to the bacterium, the active repressor produced by the regulator forms an inactive dimer with lactose (acts as an inducer).
As a result, the inactive dimer cannot bind to the operator and the three contiguous structural genes are transcribed into a polycistronic or polygenic mRNA which is then translated into three proteins (enzymes). In the absence of lactose (the inducer), the product of regulator enzyme activates inhibitor dimer that, binds to the operator and prevents transcription.
